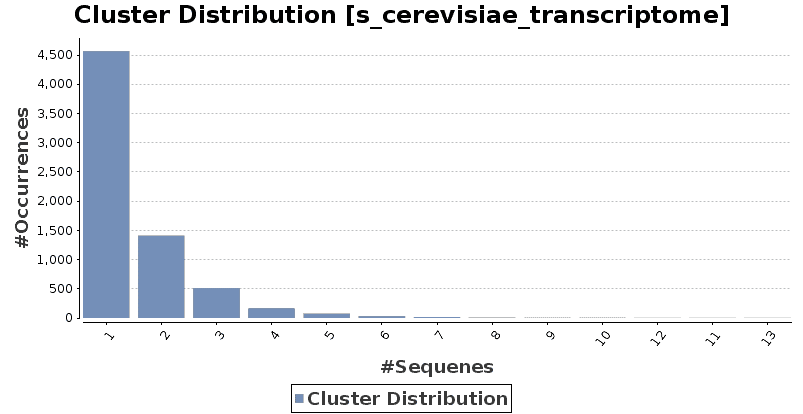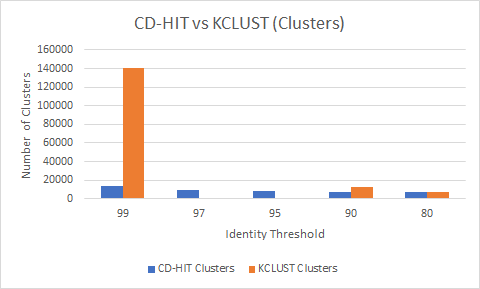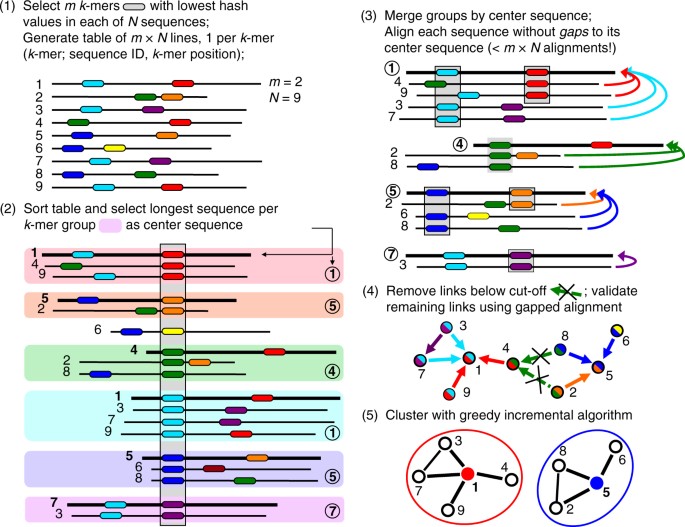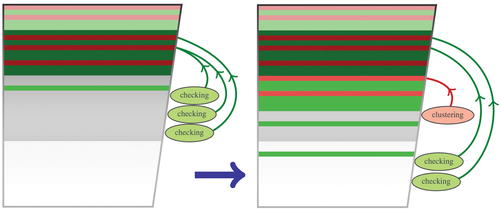
Fast Program for Clustering and Comparing Large Sets of Protein or Nucleotide Sequences | SpringerLink
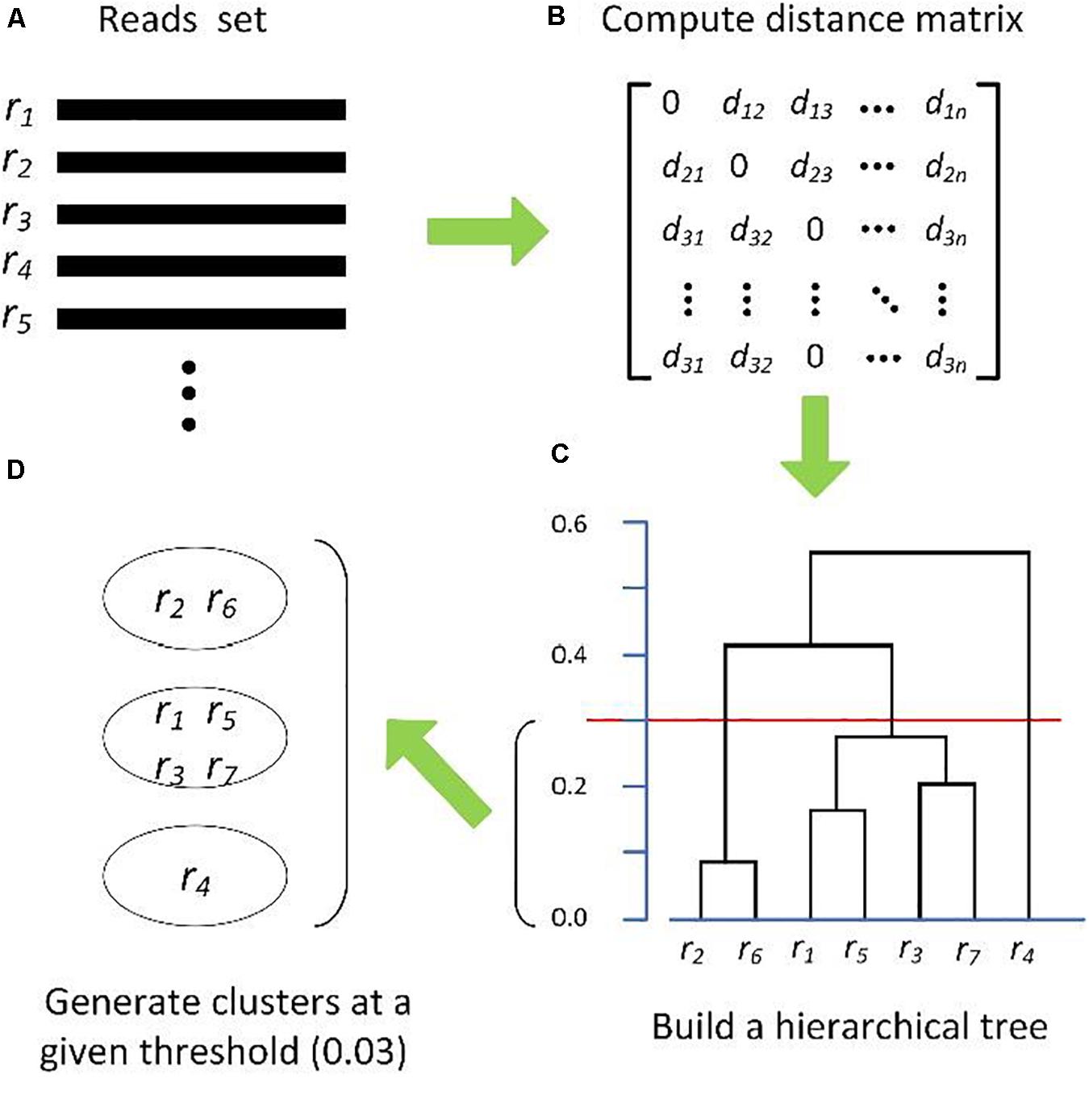
Frontiers | Comparison of Methods for Picking the Operational Taxonomic Units From Amplicon Sequences | Microbiology
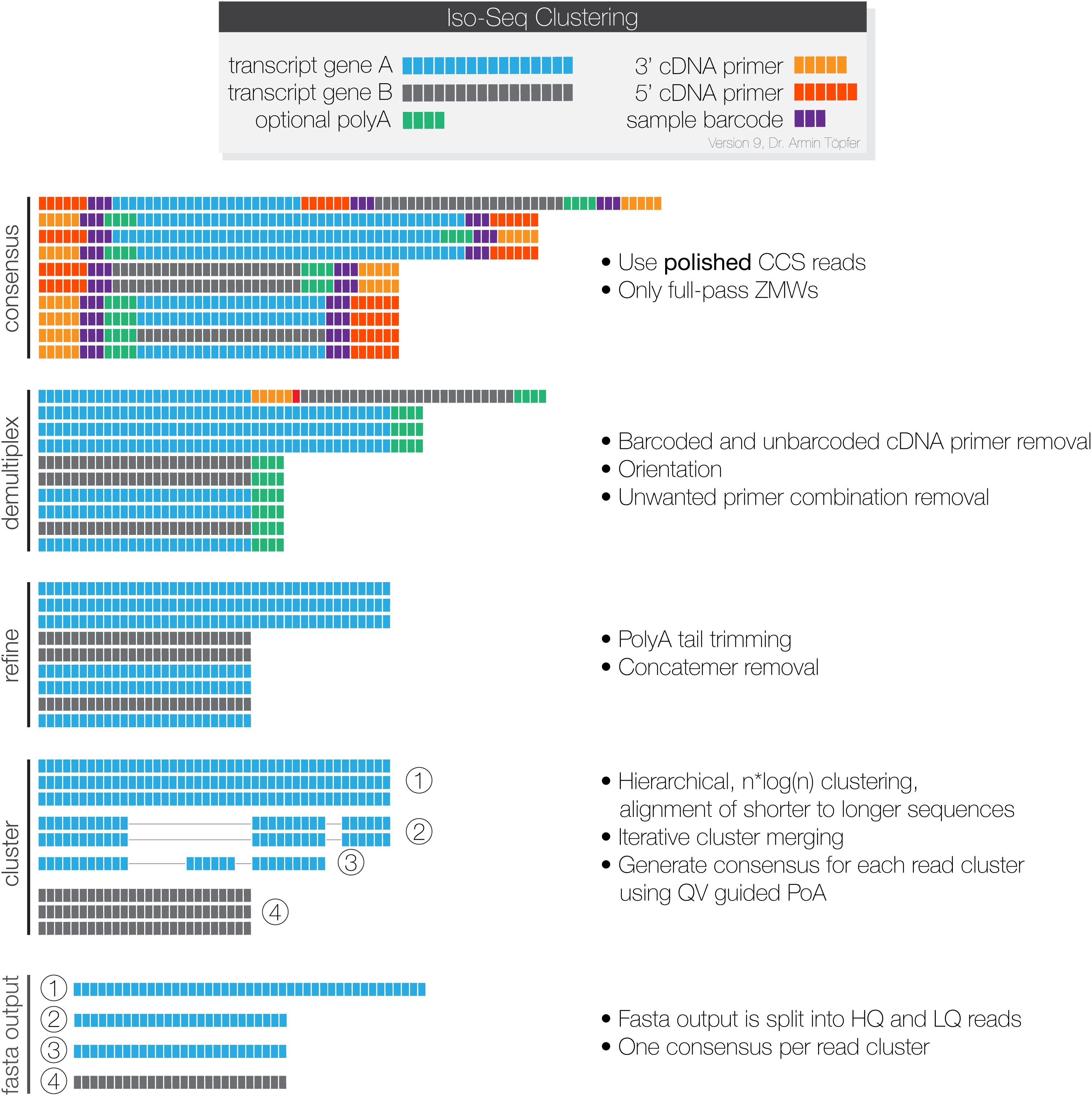
Frontiers | A de novo Full-Length mRNA Transcriptome Generated From Hybrid-Corrected PacBio Long-Reads Improves the Transcript Annotation and Identifies Thousands of Novel Splice Variants in Atlantic Salmon | Genetics
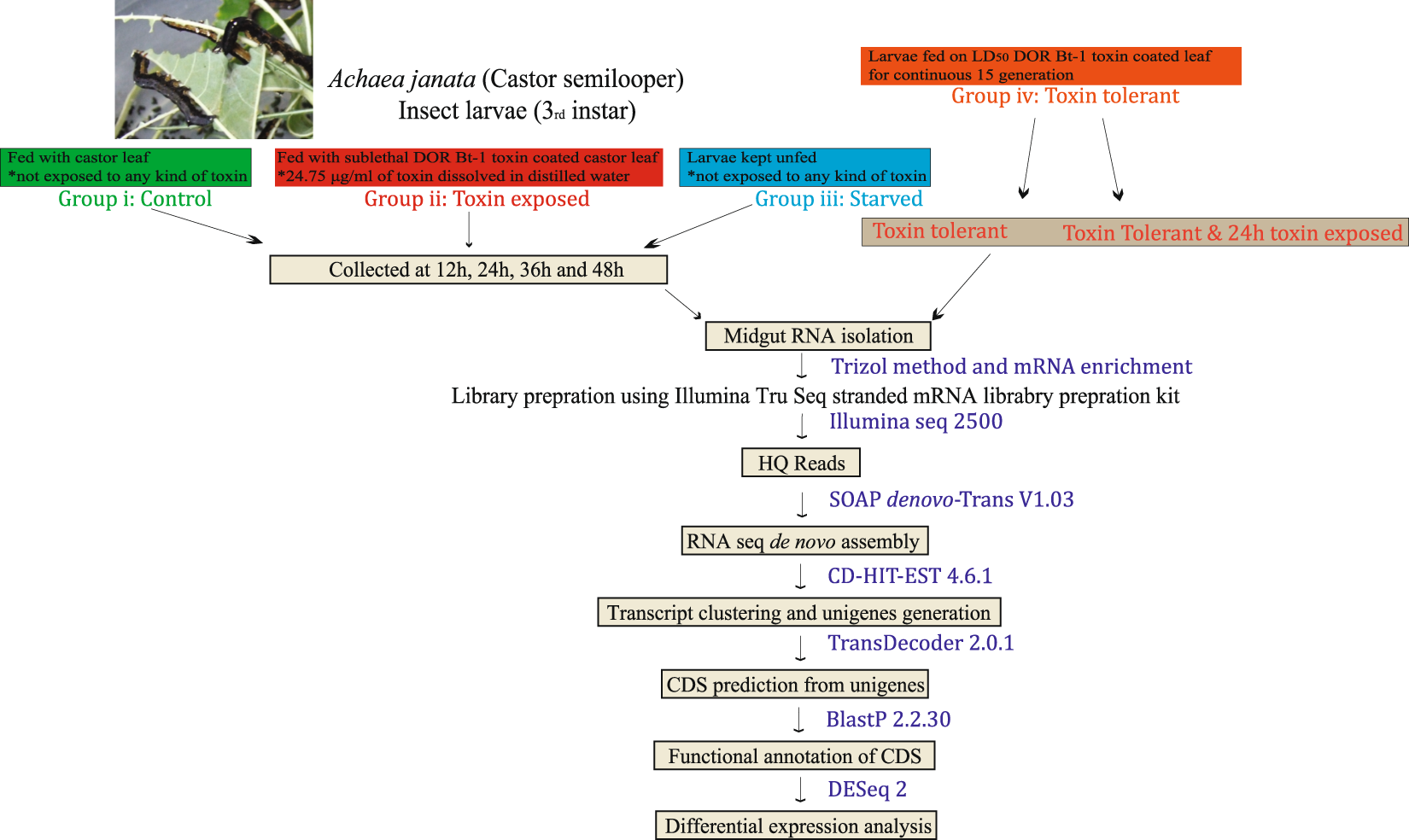
RNA-Seq analysis and de novo transcriptome assembly of Cry toxin susceptible and tolerant Achaea janata larvae | Scientific Data
CD-HIT: accelerated for clustering the next-generation sequencing data | Bioinformatics | Oxford Academic
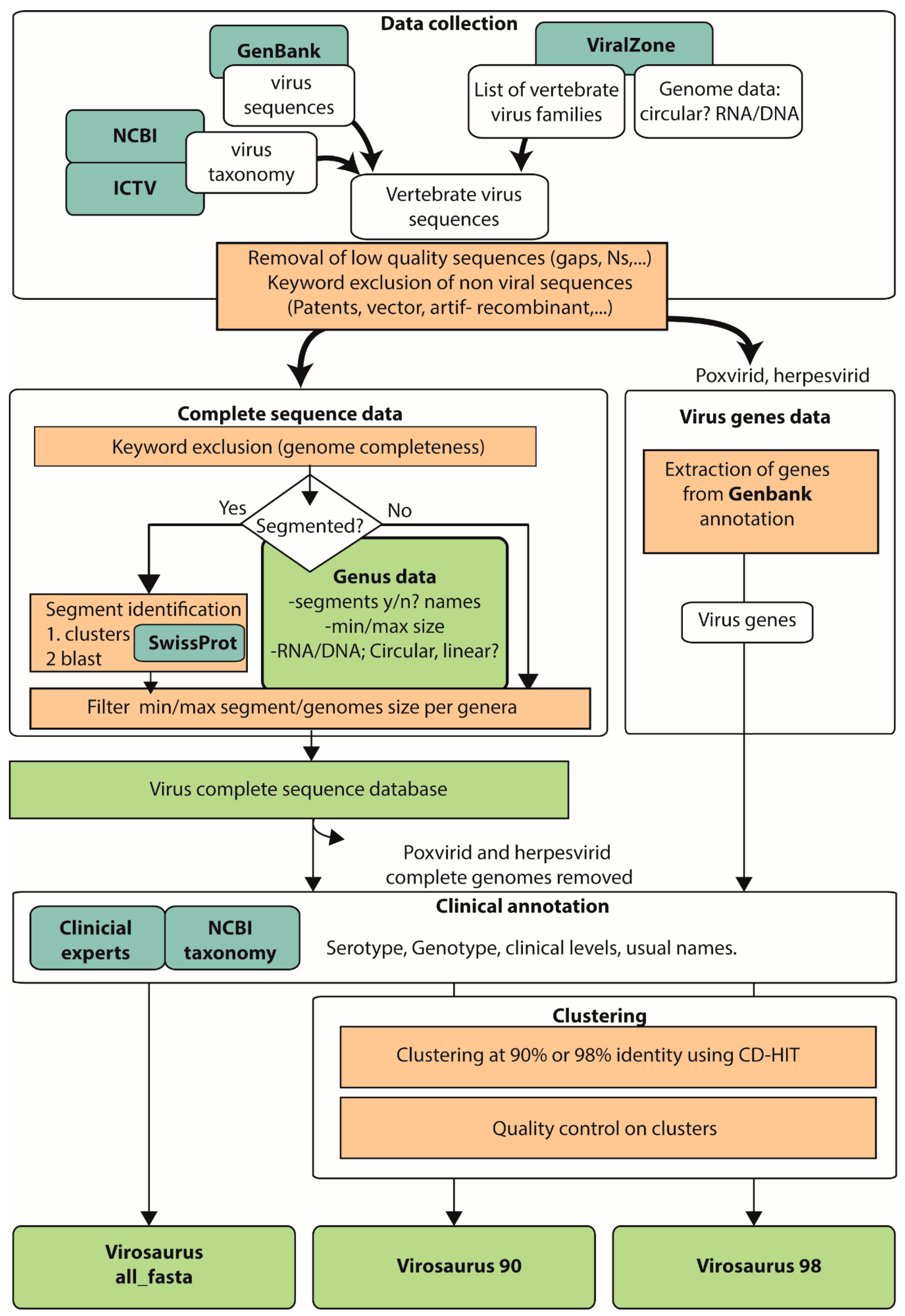
Viruses | Free Full-Text | Virosaurus A Reference to Explore and Capture Virus Genetic Diversity | HTML
MCRL: using a reference library to compress a metagenome into a non-redundant list of sequences, considering viruses as a case s
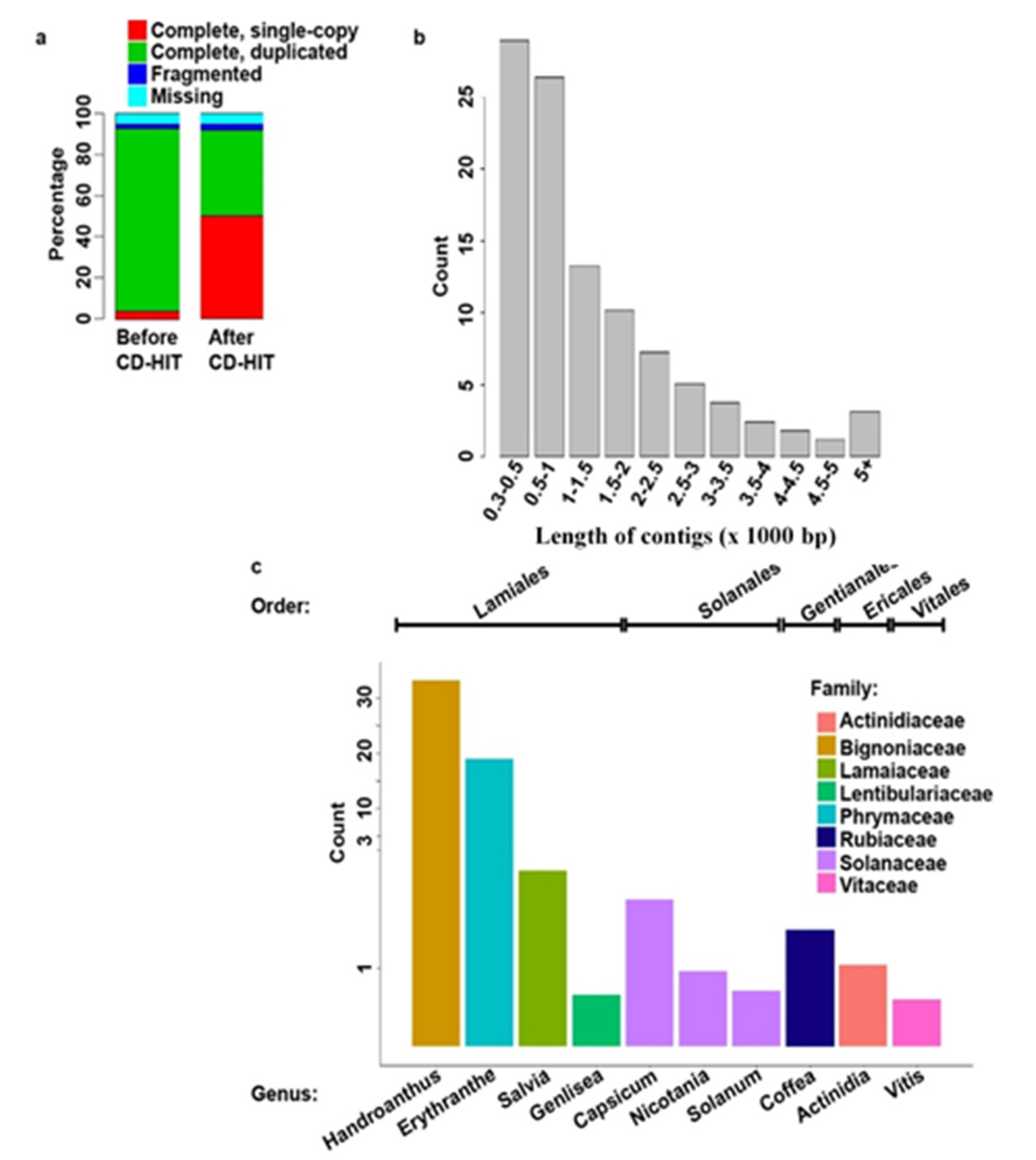
Plants | Free Full-Text | De novo Sequencing and Analysis of Salvia hispanica Tissue-Specific Transcriptome and Identification of Genes Involved in Terpenoid Biosynthesis | HTML

Fast Program for Clustering and Comparing Large Sets of Protein or Nucleotide Sequences | SpringerLink
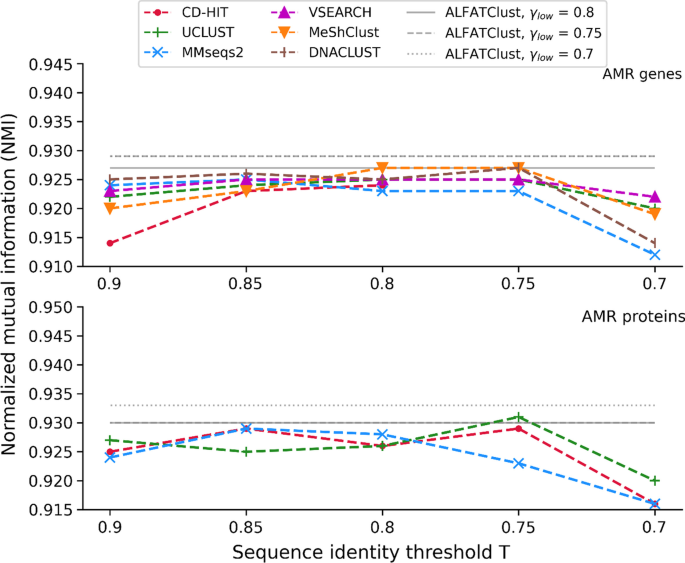
Clustering biological sequences with dynamic sequence similarity threshold | BMC Bioinformatics | Full Text
![Evaluation of full-length nanopore 16S sequencing for detection of pathogens in microbial keratitis [PeerJ] Evaluation of full-length nanopore 16S sequencing for detection of pathogens in microbial keratitis [PeerJ]](https://dfzljdn9uc3pi.cloudfront.net/2021/10778/1/fig-1-full.png)


![PDF] CD-HIT: accelerated for clustering the next-generation sequencing data | Semantic Scholar PDF] CD-HIT: accelerated for clustering the next-generation sequencing data | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/43a15ba37c0e1c88ebf28ff7f5cbe7e4ad20d6cf/2-Table1-1.png)